Context
Currently, all new drugs need to be assessed for cardiotoxicity. This normally takes place in the preclinical stages of drug development and formulation of these medications.
Regulatory bodies that approve medicines are now including in-silico cardiotoxicity in the safety assessment which employ mathematical models of cardiomyocytes. This approach means that drug safety testing does not need to rely on animal testing – which was the traditional way to test new medicines. This project therefore aligns with the 3Rs principles – Replacement, Reduction, and Refinement – aimed at enhancing the ethical treatment of animals in research.
By providing early insights into the effects of drugs within a human population, this research could speed up drug development while simultaneously reducing costs. Furthermore, this would reduce the need for animal research in drug development procedures.
Project Goals
The aim of this project is to consider the complexity of the heart to reproduce data directly comparable to measures from humans enrolled in clinical trials. As such, there are a large number of variables that need to be assessed to reproduce a human clinical trial. The current data models provide markers that correspond to clinically observed risks of heart arrhythmias, but often fail to reproduce data that can be directly compared to similar clinical measurements.
An essential stride in this project involves crafting a control population, designed to incorporate different observed effects such as placebo effects and adverse effects usually observed in healthy male and female subjects.
Rigorous validation of the computational framework is required to emulate human physiology and testing under diverse scenarios and thus ensure an accurate and reliable data set that will eventually get regulatory acceptance of the medicines before being able to market them. This is achieved through the development of computational models of electrophysiology observed in a human heart.
This project is part of a long-term research area. Currently, the inclusion of a population that includes pathophysiological states is of high importance for the extension of the computational framework, not only to reproduce the normal response to drug administration, but to stratify the use of drugs according to known pathophysiology of patients before a drug is administered.
The framework's extension also encompasses diverse life stages such as the elderly, post-menopausal, and pregnant women. This strategic expansion aims to facilitate patient-specific drug prescriptions, thereby revolutionizing drug administration protocols.
Research Team
Given the significance of this topic, numerous research groups are interested in this project, which is currently led by the Barcelona Supercomputing Center (BSC) with one Principal Investigator and three PhD students. The COVID-19 pandemic helped to accelerate preliminary work on the methodology, which was used to assess the potential proarrhythmic risk observed in COVID19 patients, after the administration of certain auto-immune and antibiotic medications.
Technical Challenges
Reproducing human physiology requires the creation of a diverse set of data models that represent a human population. With limited data available on normal human physiology, assumptions based on scientific data and medical experience are made to reproduce the physiology in a normal healthy population and predict the safety response to drug administration. Part of the challenge is to create a large set of data able to replicate sensitivity and uncertainty quantification studies in order to assess that the set of assumptions made are confirmed in the physiological responses of this population to the medicine.
Computational Methods
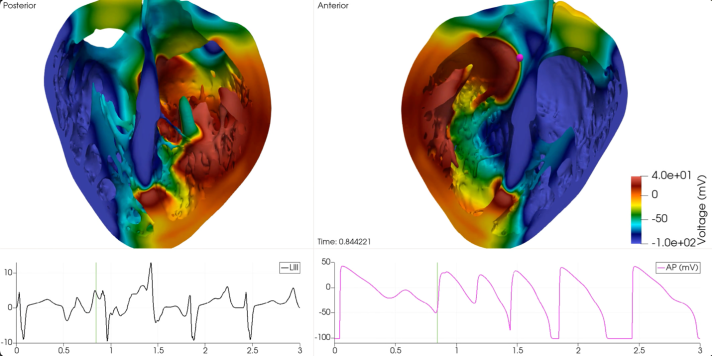
The core computational methodology of this research rests upon a computational infrastructure that combines high-resolution MRI scans of human heart donors with mathematical modeling. The output of this replicates the anatomy of the human heart. At its core is the human cardiomyocyte model which is replicated using a system of ordinary and partial differential equations.
This mathematical framework replicates the electrochemical processes occurring in cardiac tissue, resulting in the calculation of a pseudo-electrocardiogram. This is then correlated with clinical electrocardiographic markers such as QT-interval prolongation marker, which is widely regarded as the gold standard for evaluating the cardiac safety of drugs.
The time allocated on the VEGA supercomputer has played a crucial role in the production of this project. The HPC system simulated a complete human clinical safety trial which was for validating the methodology used to assess the cardiac safety of drugs.
The full production of a human clinical safety trial to assess the cardiac safety of drugs, included a control population with a placebo effect. This would have been impossible to conduct without the use of EuroHPC supercomputer’s capabilities. An integral aspect of this study is the extensive use of the Alya code, which was developed by the Barcelona Supercomputing Center (BSC). As part of the United European Application Benchmark Suite (UEABS), Alya has been tested and used for production on the EuroHPC supercomputers among others, showing scalability up to 100k CPUs in Blue Water.
Future Utilisation of EuroHPC Supercomputing Resources
EuroHPC resources have been essential to the research and the Principal Investigator intends to apply for future projects to employ EuroHPC resources. Much work is still required to reproduce human physiology and pathophysiology and validate this computational framework for potential clinical, industrial and academic use.